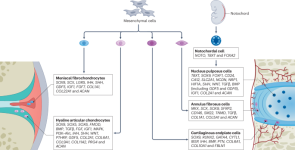
Insights into chondrocyte populations in cartilaginous tissues at the single-cell level
Liu, S. et al. Global burden of musculoskeletal disorders and attributable factors in 204 countries and territories: a secondary analysis of the global burden of disease 2019 study. BMJ Open 12, e062183 (2022).
Nguyen, A. T. et al. Musculoskeletal health: an ecological study assessing disease burden and research funding. Lancet Reg. Health Am. 29, 100661 (2024).
Zhang, Y. & Zhou, Y. Advances in targeted therapies for age-related osteoarthritis: a comprehensive review of current research. Biomed. Pharmacother. 179, 117314 (2024).
Archer, C. W. & Francis-West, P. The chondrocyte. Int. J. Biochem. Cell Biol. 35, 401–404 (2003).
Goldring, M. B. Update on the biology of the chondrocyte and new approaches to treating cartilage diseases. Best. Pract. Res. Clin. Rheumatol. 20, 1003–1025 (2006).
Muir, H. The chondrocyte, architect of cartilage. Biomechanics, structure, function and molecular biology of cartilage matrix macromolecules. Bioessays 17, 1039–1048 (1995).
Pretemer, Y. et al. Differentiation of hypertrophic chondrocytes from human iPSCs for the in vitro modeling of chondrodysplasias. Stem Cell Rep. 16, 610–625 (2021).
Riegger, J. & Brenner, R. E. Pathomechanisms of posttraumatic osteoarthritis: chondrocyte behavior and fate in a precarious environment. Int. J. Mol. Sci. 21, 1560 (2020).
Cancedda, R., Castagnola, P., Cancedda, F. D., Dozin, B. & Quarto, R. Developmental control of chondrogenesis and osteogenesis. Int. J. Dev. Biol. 44, 707–714 (2000).
Styczynska-Soczka, K., Amin, A. K. & Hall, A. C. Cell-associated type I collagen in nondegenerate and degenerate human articular cartilage. J. Cell Physiol. 236, 7672–7681 (2021).
Murray, D. H., Bush, P. G., Brenkel, I. J. & Hall, A. C. Abnormal human chondrocyte morphology is related to increased levels of cell-associated IL-1β and disruption to pericellular collagen type VI. J. Orthop. Res. 28, 1507–1514 (2010).
Hall, A. C. The role of chondrocyte morphology and volume in controlling phenotype — implications for osteoarthritis, cartilage repair, and cartilage engineering. Curr. Rheumatol. Rep. 21, 38 (2019).
Wang, T. et al. Single-cell RNA sequencing in orthopedic research. Bone Res. 11, 10 (2023).
Huang, C. et al. Single-cell transcriptomic analysis of chondrocytes in cartilage and pathogenesis of osteoarthritis. Genes. Dis. 12, 101241 (2025).
Pandey, A. & Bhutani, N. Profiling joint tissues at single-cell resolution: advances and insights. Nat. Rev. Rheumatol. 20, 7–20 (2024).
Liu, Z. et al. Single-cell profiling uncovers synovial fibroblast subpopulations associated with chondrocyte injury in osteoarthritis. Front. Endocrinol. 15, 1479909 (2024).
Danalache, M. et al. Exploration of changes in spatial chondrocyte organisation in human osteoarthritic cartilage by means of 3D imaging. Sci. Rep. 11, 9783 (2021).
Dominici, M. et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8, 315–317 (2006).
Campbell, D. D. & Pei, M. Surface markers for chondrogenic determination: a highlight of synovium-derived stem cells. Cells 1, 1107–1120 (2012).
Studle, C. et al. Challenges toward the identification of predictive markers for human mesenchymal stromal cells chondrogenic potential. Stem Cell Transl. Med. 8, 194–204 (2019).
Sober, S. A., Darmani, H., Alhattab, D. & Awidi, A. Flow cytometric characterization of cell surface markers to differentiate between fibroblasts and mesenchymal stem cells of different origin. Arch. Med. Sci. 19, 1487–1496 (2023).
Sandell, L. J. & Aigner, T. Articular cartilage and changes in arthritis. An introduction: cell biology of osteoarthritis. Arthritis Res. 3, 107–113 (2001).
Chen, N., Wu, R. W. H., Lam, Y., Chan, W. C. W. & Chan, D. Hypertrophic chondrocytes at the junction of musculoskeletal structures. Bone Rep. 19, 101698 (2023).
Schumacher, B. L., Hughes, C. E., Kuettner, K. E., Caterson, B. & Aydelotte, M. B. Immunodetection and partial cDNA sequence of the proteoglycan, superficial zone protein, synthesized by cells lining synovial joints. J. Orthop. Res. 17, 110–120 (1999).
Sun, Z. et al. Single-cell RNA sequencing reveals different chondrocyte states in femoral cartilage between osteoarthritis and healthy individuals. Front. Immunol. 15, 1407679 (2024).
Fickert, S., Fiedler, J. & Brenner, R. E. Identification of subpopulations with characteristics of mesenchymal progenitor cells from human osteoarthritic cartilage using triple staining for cell surface markers. Arthritis Res. Ther. 6, R422–432 (2004).
Tallheden, T. et al. Phenotypic plasticity of human articular chondrocytes. J. Bone Jt Surg. Am. 85, 93–100 (2003).
Jiang, Y. & Tuan, R. S. Origin and function of cartilage stem/progenitor cells in osteoarthritis. Nat. Rev. Rheumatol. 11, 206–212 (2015).
Herren, A. O. F., Amin, A. K. & Hall, A. C. A disintegrin and metalloproteinase with thrombospondin motifs-4 levels in chondrocytes of different morphology within nondegenerate and early osteoarthritic human femoral head cartilage. Cartilage 15, 278–282 (2024).
Lauer, J. C., Selig, M., Hart, M. L., Kurz, B. & Rolauffs, B. Articular chondrocyte phenotype regulation through the cytoskeleton and the signaling processes that originate from or converge on the cytoskeleton: towards a novel understanding of the intersection between actin dynamics and chondrogenic function. Int. J. Mol. Sci. 22, 3279 (2021).
Gao, H. et al. Identification of chondrocyte subpopulations in osteoarthritis using single-cell sequencing analysis. Gene 852, 147063 (2023).
Chou, C. H. et al. Synovial cell cross-talk with cartilage plays a major role in the pathogenesis of osteoarthritis. Sci. Rep. 10, 10868 (2020).
Ji, Q. et al. Single-cell RNA-seq analysis reveals the progression of human osteoarthritis. Ann. Rheum. Dis. 78, 100–110 (2019).
Di, J. et al. Cartilage tissue from sites of weight bearing in patients with osteoarthritis exhibits a differential phenotype with distinct chondrocytes subsets. RMD Open 9, e003255 (2023).
Yan, M. et al. Single-cell RNA sequencing reveals distinct chondrocyte states in femoral cartilage under weight-bearing load in rheumatoid arthritis. Front. Immunol. 14, 1247355 (2023).
Wang, X. et al. Comparison of the major cell populations among osteoarthritis, Kashin-Beck disease and healthy chondrocytes by single-cell RNA-seq analysis. Cell Death Dis. 12, 551 (2021).
Fan, Y. et al. Unveiling inflammatory and prehypertrophic cell populations as key contributors to knee cartilage degeneration in osteoarthritis using multi-omics data integration. Ann. Rheum. Dis. 83, 926–944 (2024).
Wang, J. et al. Single-cell RNA sequencing reveals the impact of mechanical loading on knee tibial cartilage in osteoarthritis. Int. Immunopharmacol. 128, 111496 (2024).
Qu, Y. et al. A comprehensive analysis of single-cell RNA transcriptome reveals unique SPP1+ chondrocytes in human osteoarthritis. Comput. Biol. Med. 160, 106926 (2023).
Liu, Y. et al. Single-cell transcriptomics reveals novel chondrocyte and osteoblast subtypes and their role in knee osteoarthritis pathogenesis. Signal. Transduct. Target. Ther. 10, 40 (2025).
Guang, Z., Min, Z., Jun-Tan, L., Tian-Xu, D. & Xiang, G. Single-cell protein activity analysis reveals a novel subpopulation of chondrocytes and the corresponding key master regulator proteins associated with anti-senescence and OA progression. Front. Immunol. 14, 1077003 (2023).
Grandi, F. C. et al. Single-cell mass cytometry reveals cross-talk between inflammation-dampening and inflammation-amplifying cells in osteoarthritic cartilage. Sci. Adv. 6, eaay5352 (2020).
Sahu, N., Grandi, F. C. & Bhutani, N. A single-cell mass cytometry platform to map the effects of preclinical drugs on cartilage homeostasis. JCI Insight 7, e160702 (2022).
Day, B., Mackenzie, W. G., Shim, S. S. & Leung, G. The vascular and nerve supply of the human meniscus. Arthroscopy 1, 58–62 (1985).
Brophy, R. H. et al. Transcriptome comparison of meniscus from patients with and without osteoarthritis. Osteoarthritis Cartilage 26, 422–432 (2018).
Jiang, Z. et al. Whole-transcriptome sequence of degenerative meniscus cells unveiling diagnostic markers and therapeutic targets for osteoarthritis. Front. Genet. 12, 754421 (2021).
Englund, M., Guermazi, A. & Lohmander, S. L. The role of the meniscus in knee osteoarthritis: a cause or consequence? Radiol. Clin. North. Am. 47, 703–712 (2009).
Fu, W. et al. Cellular features of localized microenvironments in human meniscal degeneration: a single-cell transcriptomic study. eLife 11, e79585 (2022).
Sun, H. et al. Single-cell RNA-seq analysis identifies meniscus progenitors and reveals the progression of meniscus degeneration. Ann. Rheum. Dis. 79, 408–417 (2020).
Korpershoek, J. V. et al. Selection of highly proliferative and multipotent meniscus progenitors through differential adhesion to fibronectin: a novel approach in meniscus tissue engineering. Int. J. Mol. Sci. 22, 8614 (2021).
Pattappa, G. et al. Fibronectin adherent cell populations derived from avascular and vascular regions of the meniscus have enhanced clonogenicity and differentiation potential under physioxia. Front. Bioeng. Biotechnol. 9, 789621 (2021).
Muhammad, H. et al. Human migratory meniscus progenitor cells are controlled via the TGF-β pathway. Stem Cell Rep. 3, 789–803 (2014).
Mauck, R. L., Martinez-Diaz, G. J., Yuan, X. & Tuan, R. S. Regional multilineage differentiation potential of meniscal fibrochondrocytes: implications for meniscus repair. Anat. Rec. 290, 48–58 (2007).
Elayyan, J. et al. Lef1 ablation alleviates cartilage mineralization following posttraumatic osteoarthritis induction. Proc. Natl Acad. Sci. USA 119, e2116855119 (2022).
Rajasekaran, S. et al. Profiling extra cellular matrix associated proteome of human fetal nucleus pulposus in search for regenerative targets. Sci. Rep. 11, 19013 (2021).
Rajasekaran, S. et al. Proteomic signature of nucleus pulposus in fetal intervertebral disc. Asian Spine J. 14, 409–420 (2020).
Diaz-Hernandez, M. E. et al. Derivation of notochordal cells from human embryonic stem cells reveals unique regulatory networks by single cell-transcriptomics. J. Cell Physiol. 235, 5241–5255 (2020).
Rodrigues-Pinto, R. et al. Human notochordal cell transcriptome unveils potential regulators of cell function in the developing intervertebral disc. Sci. Rep. 8, 12866 (2018).
Tam, V. et al. DIPPER, a spatiotemporal proteomics atlas of human intervertebral discs for exploring ageing and degeneration dynamics. eLife 9, e64940 (2020).
Minogue, B. M., Richardson, S. M., Zeef, L. A., Freemont, A. J. & Hoyland, J. A. Characterization of the human nucleus pulposus cell phenotype and evaluation of novel marker gene expression to define adult stem cell differentiation. Arthritis Rheum. 62, 3695–3705 (2010).
Yee, A. et al. Fibrotic-like changes in degenerate human intervertebral discs revealed by quantitative proteomic analysis. Osteoarthritis Cartilage 24, 503–513 (2016).
Ye, D. et al. Comparative and quantitative proteomic analysis of normal and degenerated human annulus fibrosus cells. Clin. Exp. Pharmacol. Physiol. 42, 530–536 (2015).
Qiu, C. et al. Differential proteomic analysis of fetal and geriatric lumbar nucleus pulposus: immunoinflammation and age-related intervertebral disc degeneration. BMC Musculoskelet. Disord. 21, 339 (2020).
Riester, S. M. et al. RNA sequencing identifies gene regulatory networks controlling extracellular matrix synthesis in intervertebral disk tissues. J. Orthop. Res. 36, 1356–1369 (2018).
Risbud, M. V. et al. Defining the phenotype of young healthy nucleus pulposus cells: recommendations of the spine research interest group at the 2014 annual ORS meeting. J. Orthop. Res. 33, 283–293 (2015).
Richardson, S. M. et al. Notochordal and nucleus pulposus marker expression is maintained by sub-populations of adult human nucleus pulposus cells through aging and degeneration. Sci. Rep. 7, 1501 (2017).
Minogue, B. M., Richardson, S. M., Zeef, L. A., Freemont, A. J. & Hoyland, J. A. Transcriptional profiling of bovine intervertebral disc cells: implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res. Ther. 12, R22 (2010).
Sakai, D. et al. Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nat. Commun. 3, 1264 (2012).
Lyu, F. J. et al. IVD progenitor cells: a new horizon for understanding disc homeostasis and repair. Nat. Rev. Rheumatol. 15, 102–112 (2019).
Fernandes, L. M. et al. Single-cell RNA-seq identifies unique transcriptional landscapes of human nucleus pulposus and annulus fibrosus cells. Sci. Rep. 10, 15263 (2020).
Cherif, H. et al. Single-Cell RNA-seq analysis of cells from degenerating and non-degenerating intervertebral discs from the same individual reveals new biomarkers for intervertebral disc degeneration. Int. J. Mol. Sci. 23, 3993 (2022).
Wang, D. et al. Single-cell transcriptomics reveals heterogeneity and intercellular crosstalk in human intervertebral disc degeneration. iScience 26, 106692 (2023).
Guo, S. et al. Single-cell RNA-seq analysis reveals that immune cells induce human nucleus pulposus ossification and degeneration. Front. Immunol. 14, 1224627 (2023).
Li, Z. et al. Single-Cell RNA sequencing reveals the difference in human normal and degenerative nucleus pulposus tissue profiles and cellular interactions. Front. Cell Dev. Biol. 10, 910626 (2022).
Wang, H. et al. Decoding the annulus fibrosus cell atlas by scRNA-seq to develop an inducible composite hydrogel: a novel strategy for disc reconstruction. Bioact. Mater. 14, 350–363 (2022).
Ling, Z. et al. Single-cell RNA-seq analysis reveals macrophage involved in the progression of human intervertebral disc degeneration. Front. Cell Dev. Biol. 9, 833420 (2021).
Han, S. et al. Single-cell RNA sequencing of the nucleus pulposus reveals chondrocyte differentiation and regulation in intervertebral disc degeneration. Front. Cell Dev. Biol. 10, 824771 (2022).
Gan, Y. et al. Spatially defined single-cell transcriptional profiling characterizes diverse chondrocyte subtypes and nucleus pulposus progenitors in human intervertebral discs. Bone Res. 9, 37 (2021).
Zhou, T. et al. Spatiotemporal characterization of human early intervertebral disc formation at single-cell resolution. Adv. Sci. 10, e2206296 (2023).
Yang, X., Lu, Y., Zhou, H., Jiang, H. T. & Chu, L. Integrated proteome sequencing, bulk RNA sequencing and single-cell RNA sequencing to identify potential biomarkers in different grades of intervertebral disc degeneration. Front. Cell Dev. Biol. 11, 1136777 (2023).
Glyn-Jones, S. et al. Osteoarthritis. Lancet 386, 376–387 (2015).
Aigner, T. et al. Apoptotic cell death is not a widespread phenomenon in normal aging and osteoarthritis human articular knee cartilage: a study of proliferation, programmed cell death (apoptosis), and viability of chondrocytes in normal and osteoarthritic human knee cartilage. Arthritis Rheum. 44, 1304–1312 (2001).
Coryell, P. R., Diekman, B. O. & Loeser, R. F. Mechanisms and therapeutic implications of cellular senescence in osteoarthritis. Nat. Rev. Rheumatol. 17, 47–57 (2021).
Mobasheri, A., Matta, C., Zakany, R. & Musumeci, G. Chondrosenescence: definition, hallmarks and potential role in the pathogenesis of osteoarthritis. Maturitas 80, 237–244 (2015).
Campisi, J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 75, 685–705 (2013).
Gruber, H. E., Ingram, J. A., Norton, H. J. & Hanley, E. N. Jr. Senescence in cells of the aging and degenerating intervertebral disc: immunolocalization of senescence-associated beta-galactosidase in human and sand rat discs. Spine 32, 321–327 (2007).
Le Maitre, C. L., Freemont, A. J. & Hoyland, J. A. Accelerated cellular senescence in degenerate intervertebral discs: a possible role in the pathogenesis of intervertebral disc degeneration. Arthritis Res. Ther. 9, R45 (2007).
Tchkonia, T., Zhu, Y., van Deursen, J., Campisi, J. & Kirkland, J. L. Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. J. Clin. Invest. 123, 966–972 (2013).
Childs, B. G., Durik, M., Baker, D. J. & van Deursen, J. M. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat. Med. 21, 1424–1435 (2015).
Silwal, P. et al. Cellular senescence in intervertebral disc aging and degeneration: molecular mechanisms and potential therapeutic opportunities. Biomolecules 13, 686 (2023).
Vamvakas, S. S., Mavrogonatou, E. & Kletsas, D. Human nucleus pulposus intervertebral disc cells becoming senescent using different treatments exhibit a similar transcriptional profile of catabolic and inflammatory genes. Eur. Spine J. 26, 2063–2071 (2017).
Nelson, G. et al. A senescent cell bystander effect: senescence-induced senescence. Aging Cell 11, 345–349 (2012).
Aigner, T., Soder, S., Gebhard, P. M., McAlinden, A. & Haag, J. Mechanisms of disease: role of chondrocytes in the pathogenesis of osteoarthritis — structure, chaos and senescence. Nat. Clin. Pract. Rheumatol. 3, 391–399 (2007).
Franceschi, C. & Campisi, J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A Biol. Sci. Med. Sci. 69, S4–S9 (2014).
Martin, J. A., Brown, T., Heiner, A. & Buckwalter, J. A. Post-traumatic osteoarthritis: the role of accelerated chondrocyte senescence. Biorheology 41, 479–491 (2004).
Jeon, O. H. et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat. Med. 23, 775–781 (2017).
Dowthwaite, G. P. et al. The surface of articular cartilage contains a progenitor cell population. J. Cell Sci. 117, 889–897 (2004).
Diekman, B. O. et al. Expression of p16INK4a is a biomarker of chondrocyte aging but does not cause osteoarthritis. Aging Cell 17, e12771 (2018).
Rim, Y. A., Nam, Y. & Ju, J. H. The role of chondrocyte hypertrophy and senescence in osteoarthritis initiation and progression. Int. J. Mol. Sci. 21, 2358 (2020).
Langley, E. et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 21, 2383–2396 (2002).
Batshon, G. et al. Serum NT/CT SIRT1 ratio reflects early osteoarthritis and chondrosenescence. Ann. Rheum. Dis. 79, 1370–1380 (2020).
Makarczyk, M. J. et al. Aging-associated increase of GATA4 levels in articular cartilage is linked to impaired regenerative capacity of chondrocytes and osteoarthritis. Osteoarthritis Cartilage (2025).
Ma, H. L. et al. Osteoarthritis severity is sex dependent in a surgical mouse model. Osteoarthritis Cartilage 15, 695–700 (2007).
Zhra, M. et al. The expression of a subset of aging and antiaging markers following the chondrogenic and osteogenic differentiation of mesenchymal stem cells of placental origin. Cells 13, 1022 (2024).
Maatuf, Y. H. et al. Diverse response to local pharmacological blockade of Sirt1 cleavage in age-induced versus trauma-induced osteoarthritis female mice. Biomolecules 14, 81 (2024).
Tsujii, A., Nakamura, N. & Horibe, S. Age-related changes in the knee meniscus. Knee 24, 1262–1270 (2017).
Chen, M. et al. Identification and validation of pivotal genes related to age-related meniscus degeneration based on gene expression profiling analysis and in vivo and in vitro models detection. BMC Med. Genomics 14, 237 (2021).
Lee, K. I. et al. FOXO1 and FOXO3 transcription factors have unique functions in meniscus development and homeostasis during aging and osteoarthritis. Proc. Natl Acad. Sci. USA 117, 3135–3143 (2020).
Swahn, H. et al. Senescent cell population with ZEB1 transcription factor as its main regulator promotes osteoarthritis in cartilage and meniscus. Ann. Rheum. Dis. 82, 403–415 (2023).
Novais, E. J. et al. Long-term treatment with senolytic drugs dasatinib and quercetin ameliorates age-dependent intervertebral disc degeneration in mice. Nat. Commun. 12, 5213 (2021).
Novais, E. J., Diekman, B. O., Shapiro, I. M. & Risbud, M. V. p16Ink4a deletion in cells of the intervertebral disc affects their matrix homeostasis and senescence associated secretory phenotype without altering onset of senescence. Matrix Biol. 82, 54–70 (2019).
Che, H. et al. p16 deficiency attenuates intervertebral disc degeneration by adjusting oxidative stress and nucleus pulposus cell cycle. eLife 9, e52570 (2020).
Patil, P. et al. Systemic clearance of p16INK4a-positive senescent cells mitigates age-associated intervertebral disc degeneration. Aging Cell 18, e12927 (2019).
Cherif, H. et al. Senotherapeutic drugs for human intervertebral disc degeneration and low back pain. eLife 9, e54693 (2020).
Cherif, H. et al. Curcumin and o-vanillin exhibit evidence of senolytic activity in human IVD cells in vitro. J. Clin. Med. 8, 433 (2019).
Swahn, H. et al. Shared and compartment-specific processes in nucleus pulposus and annulus fibrosus during intervertebral disc degeneration. Adv. Sci. 11, e2309032 (2024).
Zhang, Y. et al. Single-cell RNA-seq analysis identifies unique chondrocyte subsets and reveals involvement of ferroptosis in human intervertebral disc degeneration. Osteoarthritis Cartilage 29, 1324–1334 (2021).
Tang, X. et al. ATG9A as a potential diagnostic marker of intervertebral disc degeneration: Inferences from experiments and bioinformatics analysis incorporating sc-RNA-seq data. Gene 897, 148084 (2024).
Madhu, V. et al. The mitophagy receptor BNIP3 is critical for the regulation of metabolic homeostasis and mitochondrial function in the nucleus pulposus cells of the intervertebral disc. Autophagy 19, 1821–1843 (2023).
Madhu, V., Guntur, A. R. & Risbud, M. V. Role of autophagy in intervertebral disc and cartilage function: implications in health and disease. Matrix Biol. 100–101, 207–220 (2021).
Lee, Y. et al. Coordinate regulation of the senescent state by selective autophagy. Dev. Cell 56, 1512–1525.e7 (2021).
Ottone, O. K., Kim, C. J., Collins, J. A. & Risbud, M. V. The cGAS-STING pathway affects vertebral bone but does not promote intervertebral disc cell senescence or degeneration. Front. Immunol. 13, 882407 (2022).
Hatami-Marbini, H. On the mechanical roles of glycosaminoglycans in the tensile properties of porcine corneal stroma. Invest. Ophthalmol. Vis. Sci. 64, 3 (2023).
Sarrigiannidis, S. O. et al. A tough act to follow: collagen hydrogel modifications to improve mechanical and growth factor loading capabilities. Mater. Today Bio 10, 100098 (2021).
Gama, C. I. et al. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat. Chem. Biol. 2, 467–473 (2006).
Vogel, C. & Marcotte, E. M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232 (2012).
Yao, Q. et al. Osteoarthritis: pathogenic signaling pathways and therapeutic targets. Signal. Transduct. Target. Ther. 8, 56 (2023).
Ferguson, G. B. et al. Mapping molecular landmarks of human skeletal ontogeny and pluripotent stem cell-derived articular chondrocytes. Nat. Commun. 9, 3634 (2018).
Sulaiman, S. Z. S. et al. Comparison of bone and articular cartilage changes in osteoarthritis: a micro-computed tomography and histological study of surgically and chemically induced osteoarthritic rabbit models. J. Orthop. Surg. Res. 16, 663 (2021).
Vincent, T. L. Mechanoflammation in osteoarthritis pathogenesis. Semin. Arthritis Rheum. 49, S36–S38 (2019).
Weber, P. et al. The collagenase-induced osteoarthritis (CIOA) model: where mechanical damage meets inflammation. Osteoarthritis Cartilage Open 6, 100539 (2024).
Colbath, A. & Haubruck, P. Closing the gap: sex-related differences in osteoarthritis and the ongoing need for translational studies. Ann. Transl. Med. 11, 339 (2023).
Sebastian, A. et al. Single-cell RNA-seq reveals transcriptomic heterogeneity and post-traumatic osteoarthritis-associated early molecular changes in mouse articular chondrocytes. Cells 10, 1462 (2021).
Dvir-Ginzberg, M., Maatuf, Y. H. & Mobasheri, A. Do we understand sex-related differences governing dimorphic disease mechanisms in preclinical animal models of osteoarthritis? Osteoarthritis Cartilage 32, 1054–1057 (2024).
Pan, B. et al. Identification of key biomarkers related to fibrocartilage chondrocytes for osteoarthritis based on bulk, single-cell transcriptomic data. Front. Immunol. 15, 1482361 (2024).
Chapman, J. H., Ghosh, D., Attari, S., Ude, C. C. & Laurencin, C. T. Animal models of osteoarthritis: updated models and outcome measures 2016-2023. Regen. Eng. Transl. Med. 10, 127–146 (2024).
Dou, H. et al. Osteoarthritis models: from animals to tissue engineering. J. Tissue Eng. 14, 20417314231172584 (2023).
Sengprasert, P., Kamenkit, O., Tanavalee, A. & Reantragoon, R. The immunological facets of chondrocytes in osteoarthritis: a narrative review. J. Rheumatol. (2023).
Aguiar, D. J., Johnson, S. L. & Oegema, T. R. Notochordal cells interact with nucleus pulposus cells: regulation of proteoglycan synthesis. Exp. Cell Res. 246, 129–137 (1999).



